Influenza is one of the world’s major uncontrolled diseases, the cause in humans of both mild annual epidemics and more severe intermittent pandemics, such as the ‘swine flu’ outbreak in 2009 and potentially H5N1 ‘bird flu’. Influenza is caused by the influenza virus, a member of the Orthomyxovirus family. In spite of their importance, many aspects of influenza virus replication are poorly understood.
The genome of the influenza virus is made of RNA. This genome serves as a template for both transcription (the production of mRNA in order to make viral proteins) and replication (the production of new progeny genomes), and both of these processes are carried out by the same viral polymerase enzyme; however, the mechanisms for these events are very different. Despite decades of intensive research, the question of how these processes are regulated remains elusive. Our research uses single-molecule fluorescent spectroscopy techniques such as fluorescence anisotropy, single-molecule FRET and total-internal reflection fluorescence (TIRF) microscopy, combined with structural modelling, to further understand the regulatory mechanisms of influenza virus replication.
Crystal structures of the influenza RNA polymerase have provided intriguing static snapshots of how the viral RNA genome is bound and copied. We have extended these studies by using solution-based smFRET to probe dynamic motions of the RNA that occur during polymerase binding and initial replication (1,2). We have shown that even in the absence of nucleotides the polymerase-bound RNA template exists in two conformations, corresponding to pre-initiation and initiation states (see figure below). The similar architecture and RNA-binding modes of the RNA polymerases of all segmented negative-strand viruses suggests a common evolutionary origin and mechanism of RNA synthesis; our results therefore have implications for understanding the mechanisms of viral replication by other pathogenic viruses.
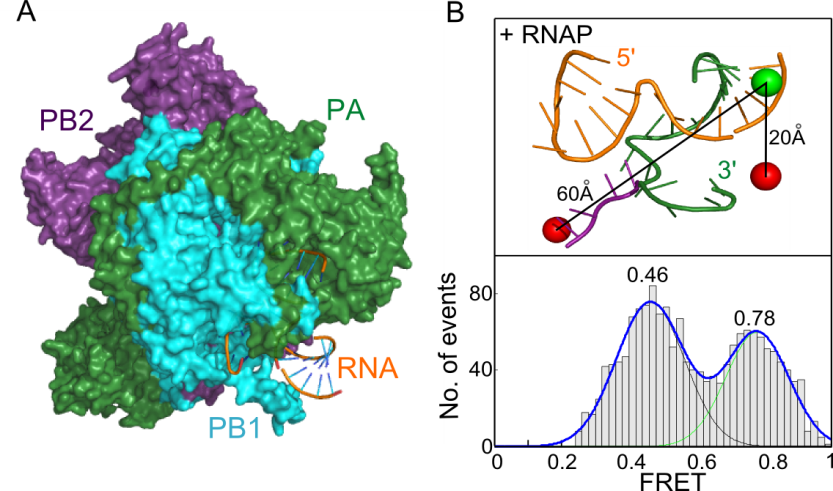 A) The 3' and 5' ends of each viral RNA segment form a partially double-stranded promoter that is bound by the viral RNA polymerase (3). B) Single-molecule FRET of diffusing RNA/RNAP complexes reveals two RNA conformations, corresponding to pre-initiation and initiation states.
A) The 3' and 5' ends of each viral RNA segment form a partially double-stranded promoter that is bound by the viral RNA polymerase (3). B) Single-molecule FRET of diffusing RNA/RNAP complexes reveals two RNA conformations, corresponding to pre-initiation and initiation states.
- Tomescu AI, Robb NC, Hengrung N, Fodor E and Kapanidis AN. Single-molecule FRET reveals a corkscrew RNA structure for the polymerase-bound influenza virus promoter. PNAS. 2014, 111(32):E3335-42.
- Robb NC, te Velthuis AJW, Wieneke R, Tampè R, Cordes T, Fodor E and Kapanidis AN. Single-molecule FRET reveals the pre-initiation and initiation conformations of influenza virus promoter RNA. NAR. 2016, 44(21):10304-10315.
- Pflug A, Guilligay D, Reich S, Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014, 516(7531):355-60.