Home
The Gene Machines group
Exploring biology, one molecule at a time
Gene expression is the vital path that converts DNA information into functional proteins. Our group studies mechanisms and machines of gene expression using single-molecule biophysical methods and biochemistry. We observe single biomachines in real time, both "in vitro" and inside living cells. Our favourite machines come from microbes, especially from the bacterium E.coli and the influenza virus.
We also develop new research tools, such as single-molecule fluorescence and super-resolution methods, microscopes, assays, and software. We then leverage our biological understanding and our new "tools" to build novel biosensors and diagnostic platforms.
Our team is a Biological Physics research group within Condensed Matter Physics, and has recently relocated to the new Oxford Kavli Institute for Nanoscience Discovery, an interdisciplinary institute focusing on using physical approaches to study cellular mechanisms. We are also linked to many of the Oxford Doctoral Training Centres (DTCs) programmes (including the Interdisciplinary Bioscience DTP and the Chemistry-in-Cells programmes).












Latest News
2026 January
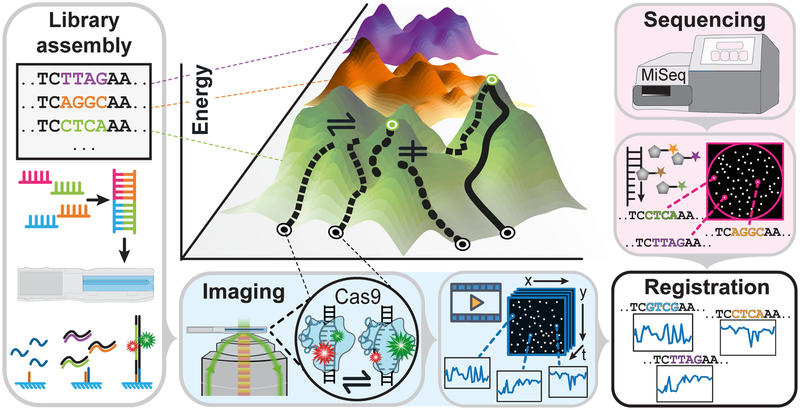
Congratulations to Achilles and Jagadish on the Science review!
This review highlights new high‑throughput single-molecule methods linking DNA/RNA sequences to their real‑time molecular behaviour, revealing how tiny changes drive big biological effects!
2025 November
Huge congratulations to Dr. Qing for acing the PhD viva—so well deserved!
2025 October
A warm welcome to Cornelius Lee! We're excited to have you on board for the scientific adventures ahead.
2025 June
Well done, Dr. Emma—congratulations on conquering your PhD viva!
2025 May
Welcome Sharika and Afroze! We’re very excited to have you on the team!
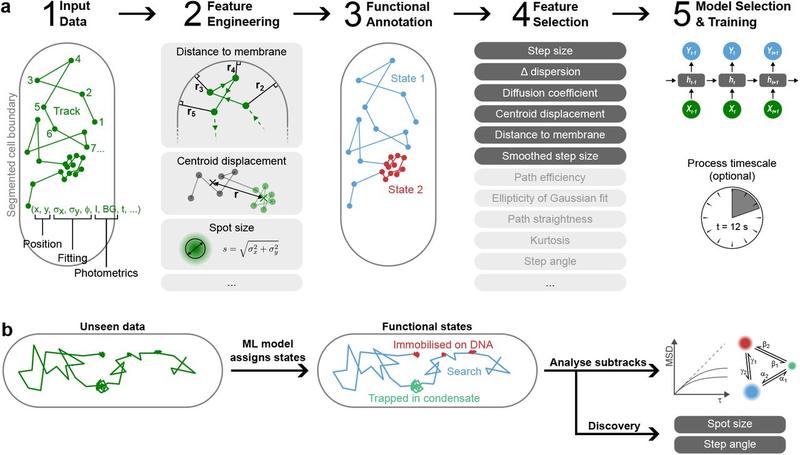
Congrats to Oli and Jacob for your new preprint!
2025 Mar
Very warm welcome to Dr. Maryam Beigzadeh to join our Lab!
2025 February
A massive congratulations to Dr. Rasched on passing the PhD viva!
2025 Jan
Congratulations Miri on passing your PhD viva!!
2024 Oct
Very warm welcome to Suchintak. We're excited to have you with us and look forward to your valuable contributions.
2024 July
It's very sad to see Piers leaving the group this month. All the best for your new addventures!
2024 Feb
Congrats to Hafez and Oli on their NusG publication, see Single-Molecule Tracking Reveals the Functional Allocation, in vivo Interactions and Spatial Organization of Universal Transcription Factor NusG
We are hiring! We have a 4-year opening for a single-molecule biophysicist AND a 4-year opening for a single-cell microbiologist. Apply by March 27th or tell your friends to do so (:-)
Our lab (Hafez, Jagadish, Oli, Achilles) is organising the 35th RNA polymerase workshop at Oxford (18-19th March, 2024). Registration is open until March 11th. Abstracts are due until March 4th. Looking fwd to hearing your new transcription and RNAP stories!
2024 Jan
Very sad to see you go, Christof! All the best for your new adventures!
2023 Dec
The infection inspection is released. Congrats to Ali on the work of Infection Inspection: Using the power of citizen science to help with image-based prediction of antibiotic resistance in Escherichia coli
2023 Nov
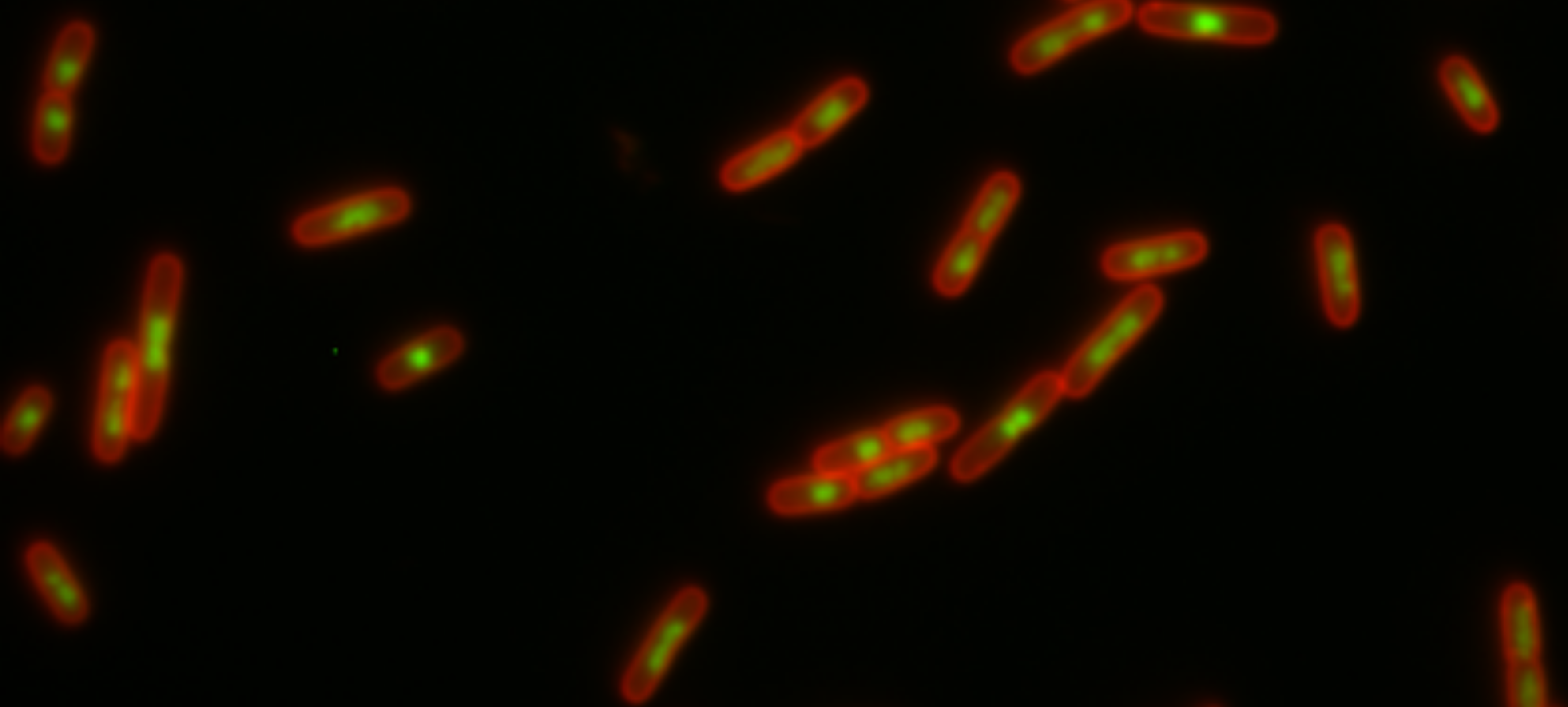
Congratulations to Alex and Piers for publishing Deep learning and single-cell phenotyping for rapid antimicrobial susceptibility detection in Escherichia coli
2023 Sep
Welcome to the lab, Ed and Jacob!
2023 Jul
Exciting preprint for Stelios! See Rapid Identification of Bacterial isolates Using Microfluidic Adaptive Channels and Multiplexed Fluorescence Microscopy
We're saying goodbye to Heesoo - best of luck with your new work back in South Korea!
Anna is sadly leaving us after finishing her PhD - she is now off to new adventures. Congratulations!
2023 Jun
Congrats to Andrew for publishing his work High-throughput super-resolution analysis of influenza virus pleomorphism reveals insights into viral spatial organization
Well done Anna for your work on The displacement of the σ70finger in initial transcription is highly heterogeneous and promoter-dependent
Congrats on finishing your PhD, Andrew! We're sad to see you go - good luck for your exciting new projects with Seamus Holden!
2023 Apr
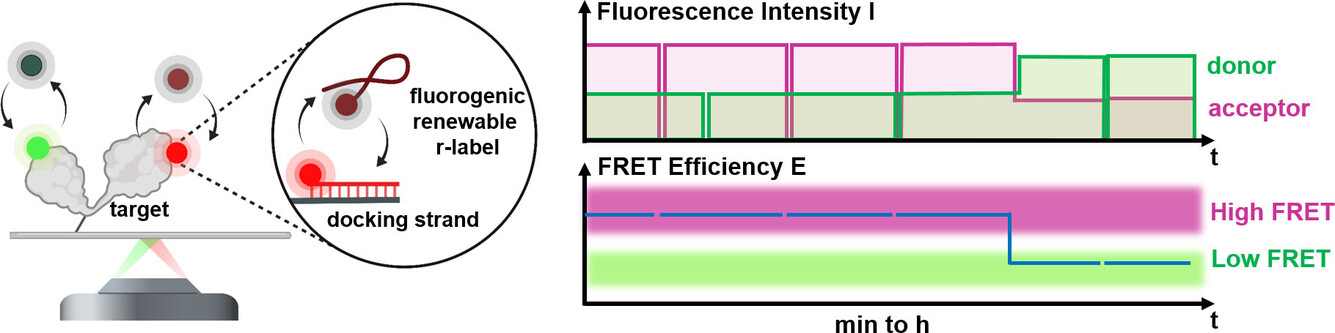
Hello REFRESH, goodbye photobleaching! Congrats Mirjam for your paper on Bleaching-resistant, Near-continuous Single-molecule Fluorescence and FRET Based on Fluorogenic and Transient DNA Binding
2022 Dec
Excited to see Nic's viral-detection paper in press! See: Virus Detection and Identification in Minutes Using Single-Particle Imaging and Deep Learning
2022 Oct
-Welcome to the Lab, Jagadish and Qing! We're excited to have you here!
2022 Feb
Congrats to Becky for the preprint of Transient DNA binding to gapped DNA substrates links DNA sequence to the single-molecule kinetics of protein-DNA interactions
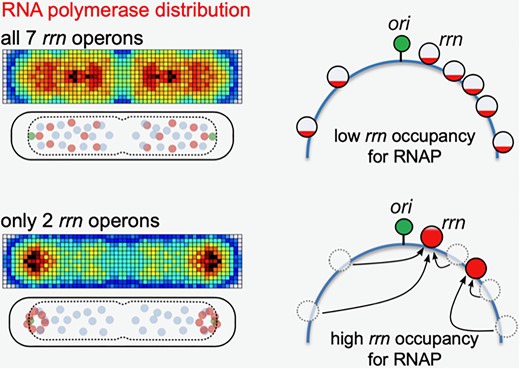
Congrats to Jun for publishing on RNA polymerase redistribution supports growth in E. coli strains with a minimal number of rRNA operons
2021 Sep
-A warm welcome to Piers! We're looking forward to working with you!


