DNA is an extremely versatile material with which to build biosensors; molecules can be encoded with binding sites specific for proteins of interest, or for other nucleic acids (DNA/RNA), and can be easily modified to contain useful chemical groups such
as fluorophores, biotin or quantum dots. Our lab have been developing biosensors for the detection of DNA-binding proteins (1-2), nucleic acids (3) and pathogens, such as viruses (3) and bacteria.
An example of a novel biosensing strategy we developed was applied to rotaviruses, the single most common cause of fatal and severe childhood diarrheal illness worldwide (>125 million cases annually). Rotavirus shares structural and functional features
with many viruses, such as the presence of segmented double-stranded RNA genomes selectively and tightly packed with a conserved number of transcription complexes in icosahedral capsids. Nascent transcripts exit the capsid through 12 channels, but
it was unknown whether these channels specialize in specific transcripts or simply act as general exit conduits. We thus developed a single-molecule assay for capturing and identifying transcripts extruded from transcriptionally active viral particles.
Our findings supported a model in which each channel specializes in extruding transcripts of a specific segment that in turn is linked to a single transcription complex. Our approach can be extended to study other viruses and transcription systems.
A further project in the lab focuses on the development of a novel biosensing assay for the rapid detection of influenza virus particles in biologically complex samples. This assay makes use of fluorescently-labelled aptamers, which are short strands
of DNA that are selected to bind with a high affinity and degree of specificity to a particular biological target.
The lab is also developing new imaging assays that contribute to the fight against the growing threat of antimicrobial resistance.
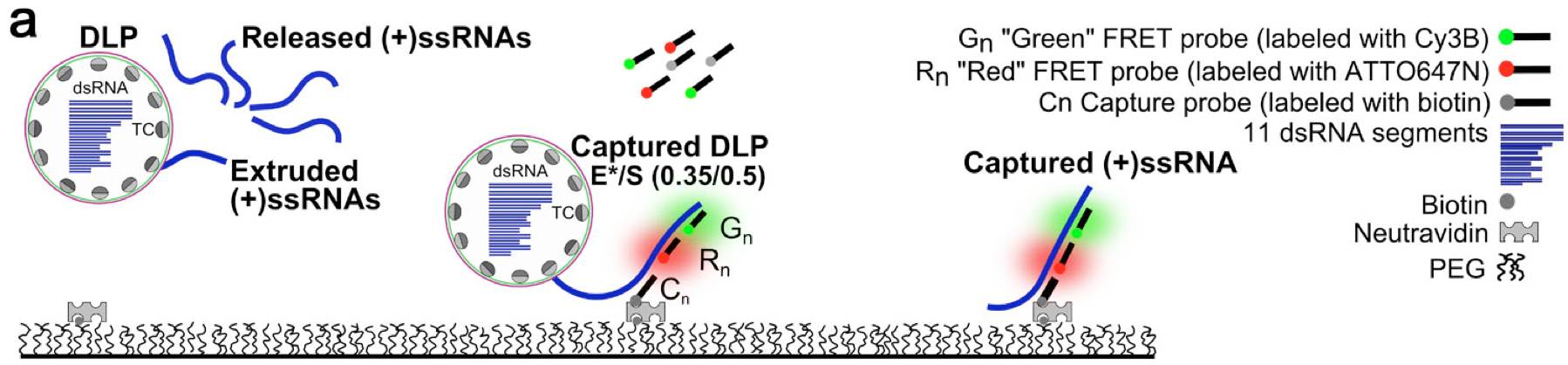 Capturing Rotavirus particles containing specific RNA prior to release, as well as specific released RNAs. Capturing is done through a biotinylated probe, whereas the identity of the RNA is determined using a single-molecule FRET signal developed between two DNA probes that hybridize with the specific Rotavirus RNA.
Capturing Rotavirus particles containing specific RNA prior to release, as well as specific released RNAs. Capturing is done through a biotinylated probe, whereas the identity of the RNA is determined using a single-molecule FRET signal developed between two DNA probes that hybridize with the specific Rotavirus RNA.
- Lymperopoulos K, Crawford R, Torella JP, Heilemann M, Hwang LC, Holden SJ, and Kapanidis AN. Single-molecule DNA biosensors for protein and ligand detection. Angew. Chem. 2010, 49 (7), pp.1316-1320.
- Crawford R, Kelly DJ, Kapanidis AN. A Protein Biosensor That Relies on Bending of Single DNA Molecules. ChemPhysChem. 2012, 13 (4), pp.918-922.
- Periz J, Celma C, Jing B, Pinkney JN, Roy P, Kapanidis AN, Rotavirus mRNAS are released by transcript-specific channels in the double-layered viral capsid, PNAS, 2013; 110, 12042–12047.